Designer Sinorhizobium meliloti strains and multi-functional vectors enable direct inter-kingdom DNA transfer
Published in PLoS One, 2019
Recommended citation: SL. Brumwell, MR. MacLeod, T. Huang, RR. Cochrane, RS. Meaney, M. Zamani, O. Matysiakiewicz, KN. Dan, P. Janakirama, DR. Edgell, TC. Charles, TM. Finan, BJ. Karas. (2019). "Designer Sinorhizobium meliloti strains and multi-functional vectors enable direct inter-kingdom DNA transfer." PLoS ONE. 14(7): e0219562. https://ryanrcochrane.github.io/files/paper1.pdf
ABSTRACT Storage, manipulation and delivery of DNA fragments is crucial for synthetic biology applications, subsequently allowing organisms of interest to be engineered with genes or pathways to produce desirable phenotypes such as disease or drought resistance in plants, or for synthesis of a specific chemical product. However, DNA with high GC content can be unstable in many host organisms including Saccharomyces cerevisiae. Here, we report the development of Sinorhizobium meliloti, a nitrogen-fixing plant symbiotic alpha-Proteobacterium, as a novel host that can store DNA, and mobilize DNA to E. coli, S. cerevisiae, and the eukaryotic microalgae Phaeodactylum tricornutum. To achieve this, we deleted the hsdR restriction-system in multiple reduced genome strains of S. meliloti that enable DNA transformation with up to 1.4 x 105 and 2.1 x 103 CFU μg-1 of DNA efficiency using electroporation and a newly developed polyethylene glycol transformation method, respectively. Multi-host and multi-functional shuttle vectors were constructed and stably propagated in S. meliloti, E. coli, S. cerevisiae, and P. tricornutum. We also developed protocols and demonstrated direct transfer of these multi-host shuttle vectors via conjugation from S. meliloti to E. coli, S. cerevisiae, and P. tricornutum. The development of S. meliloti as a new host for inter-kingdom DNA transfer will be invaluable for synthetic biology research and applications, including the installation and study of genes and biosynthetic pathways into organisms of interest in industry and agriculture.

Figures and Tables
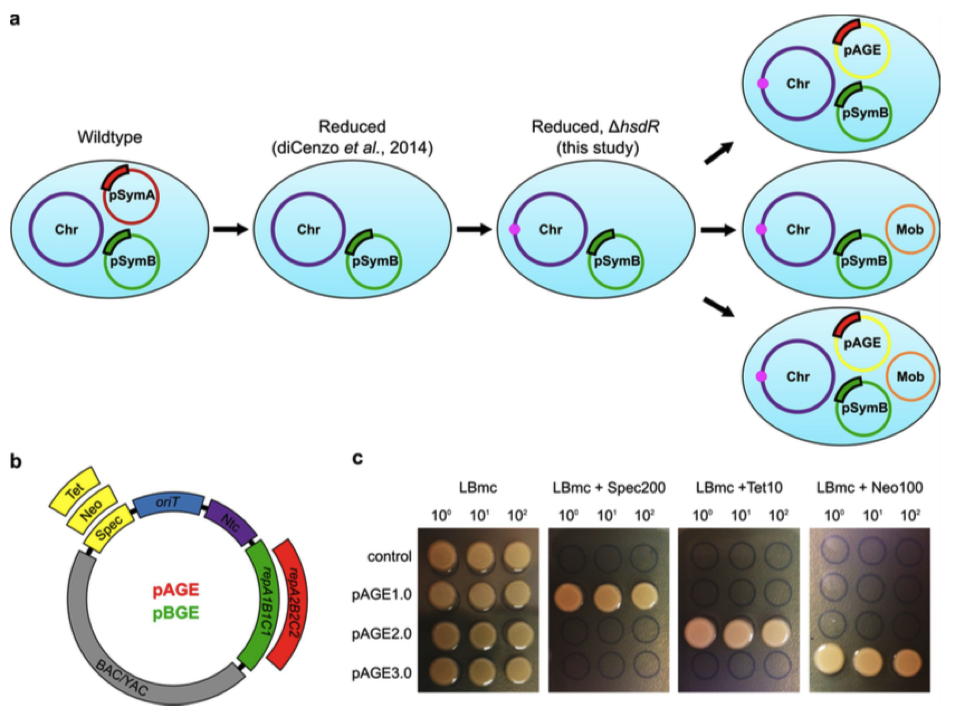 Figure 1. Development of designer S. meliloti strains and multi-functional vectors. (a) Schematic of the designer S. meliloti ΔpSymA ΔhsdR strains including the genome reduction and deletion of the restriction-system. Several versions of the designer strains were developed including strains with the pTA-Mob conjugative plasmid, and/or compatible genome engineering vectors, pAGE or pBGE, installed. (b) Vector map of pAGE/pBGE MHS vector with both standard and interchangeable components. Standard components include the BAC and YAC backbone, an origin of transfer, and the selectable marker for P. tricornutum nourseothricin N-acetyl transferase. Interchangeable components include selectable markers for S. meliloti: tetracycline, kanamycin/neomycin, and spectinomycin; and origins of replication for S. meliloti: the pSymA origin, and pSymB origin. Three vectors were constructed utilizing the repA2B2C2 origin with Spec, Tet or Neo selection, respectively. Three vectors were constructed utilizing the repA1B1C1 origin with Spec, Tet or Neo selection, respectively. (c) Following conjugation of three pAGE plasmids from E. coli ECGE101 conjugative strain to S. meliloti RmP4122 ΔpSymA ΔhsdR Nms, an antibiotic selection test for each plasmid was performed. The pAGE1.0, pAGE2.0, and pAGE3.0 vectors in S. meliloti contain antibiotic resistance markers for Spec, Tet, and Neo, respectively. S. meliloti without a plasmid was plated alongside S. meliloti harbouring pAGE1.0, pAGE2.0, or pAGE3.0 on LBmc agar, and LBmc agar supplemented with either spectinomycin, tetracycline, or neomycin.
Figure 1. Development of designer S. meliloti strains and multi-functional vectors. (a) Schematic of the designer S. meliloti ΔpSymA ΔhsdR strains including the genome reduction and deletion of the restriction-system. Several versions of the designer strains were developed including strains with the pTA-Mob conjugative plasmid, and/or compatible genome engineering vectors, pAGE or pBGE, installed. (b) Vector map of pAGE/pBGE MHS vector with both standard and interchangeable components. Standard components include the BAC and YAC backbone, an origin of transfer, and the selectable marker for P. tricornutum nourseothricin N-acetyl transferase. Interchangeable components include selectable markers for S. meliloti: tetracycline, kanamycin/neomycin, and spectinomycin; and origins of replication for S. meliloti: the pSymA origin, and pSymB origin. Three vectors were constructed utilizing the repA2B2C2 origin with Spec, Tet or Neo selection, respectively. Three vectors were constructed utilizing the repA1B1C1 origin with Spec, Tet or Neo selection, respectively. (c) Following conjugation of three pAGE plasmids from E. coli ECGE101 conjugative strain to S. meliloti RmP4122 ΔpSymA ΔhsdR Nms, an antibiotic selection test for each plasmid was performed. The pAGE1.0, pAGE2.0, and pAGE3.0 vectors in S. meliloti contain antibiotic resistance markers for Spec, Tet, and Neo, respectively. S. meliloti without a plasmid was plated alongside S. meliloti harbouring pAGE1.0, pAGE2.0, or pAGE3.0 on LBmc agar, and LBmc agar supplemented with either spectinomycin, tetracycline, or neomycin.
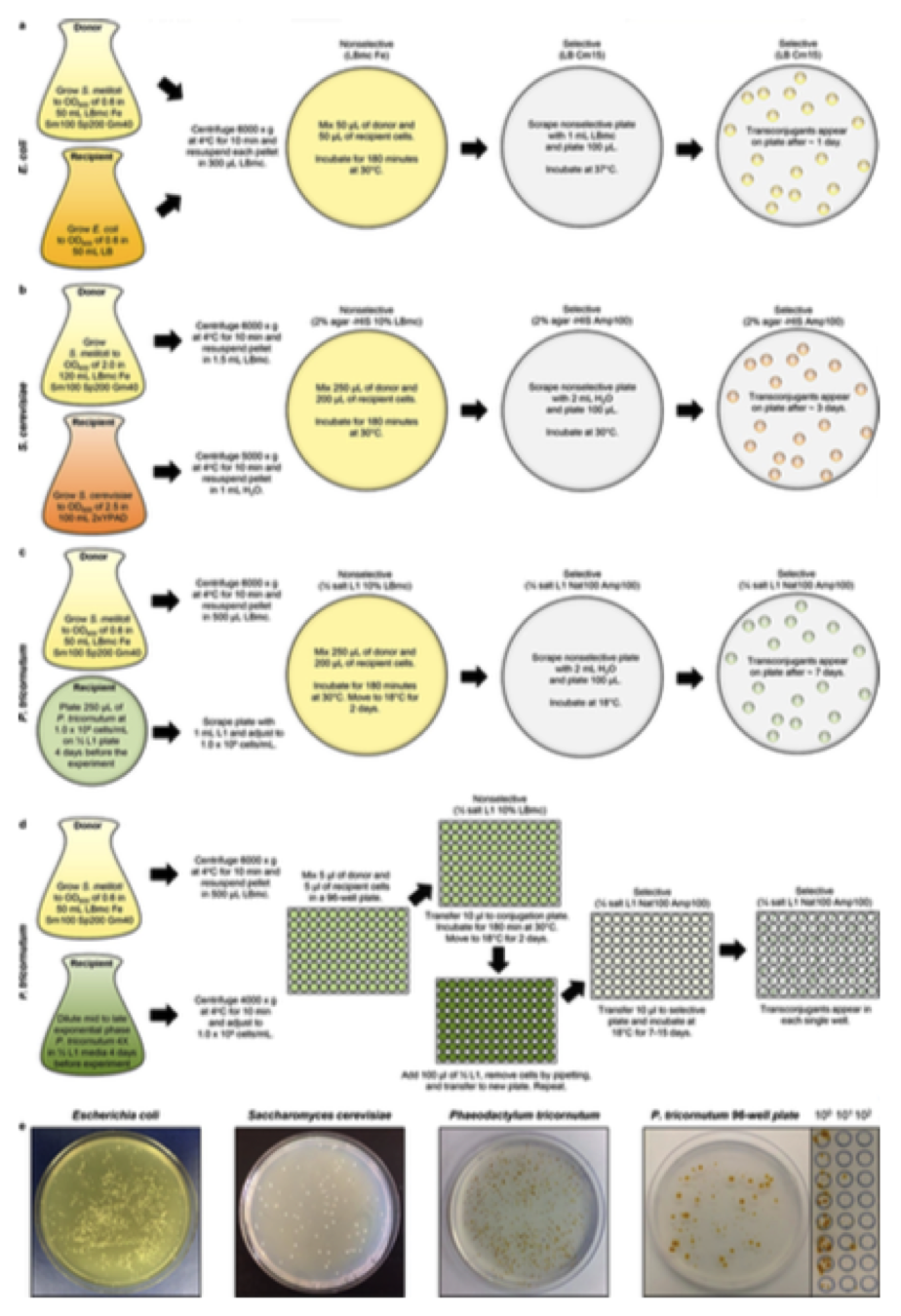 Figure 2. Optimized conjugation protocols from S. meliloti to various recipient organisms. The S. meliloti RmP4098 ΔpSymA ΔhsdR strain used is resistant to streptomycin and requires Fe supplementation for growth. The pAGE1.0 plasmid used confers resistance to spectinomycin. Media supplementation may vary with chosen S. meliloti host and MHS vector. (a) Schematic of protocol for conjugation of pAGE1.0 from S. meliloti to E. coli. (b) Schematic of protocol for conjugation of pAGE1.0 vector from S. meliloti to S. cerevisiae. (c) Schematic of protocol for conjugation of pAGE1.0 vector from S. meliloti to P. tricornutum –standard protocol. (d) Schematic of protocol for conjugation of pAGE1.0 vector from S. meliloti to P. tricornutum – 96-well plate protocol. (e) Examples of plates containing transconjugants resulting from conjugation from S. meliloti to E. coli, S. cerevisiae, and P. tricornutum. P. tricornutum transconjugants from the 96-well plate protocol are shown with 100 uL plated on a full plate, and 10 uL spot plated and serially diluted.
Figure 2. Optimized conjugation protocols from S. meliloti to various recipient organisms. The S. meliloti RmP4098 ΔpSymA ΔhsdR strain used is resistant to streptomycin and requires Fe supplementation for growth. The pAGE1.0 plasmid used confers resistance to spectinomycin. Media supplementation may vary with chosen S. meliloti host and MHS vector. (a) Schematic of protocol for conjugation of pAGE1.0 from S. meliloti to E. coli. (b) Schematic of protocol for conjugation of pAGE1.0 vector from S. meliloti to S. cerevisiae. (c) Schematic of protocol for conjugation of pAGE1.0 vector from S. meliloti to P. tricornutum –standard protocol. (d) Schematic of protocol for conjugation of pAGE1.0 vector from S. meliloti to P. tricornutum – 96-well plate protocol. (e) Examples of plates containing transconjugants resulting from conjugation from S. meliloti to E. coli, S. cerevisiae, and P. tricornutum. P. tricornutum transconjugants from the 96-well plate protocol are shown with 100 uL plated on a full plate, and 10 uL spot plated and serially diluted.
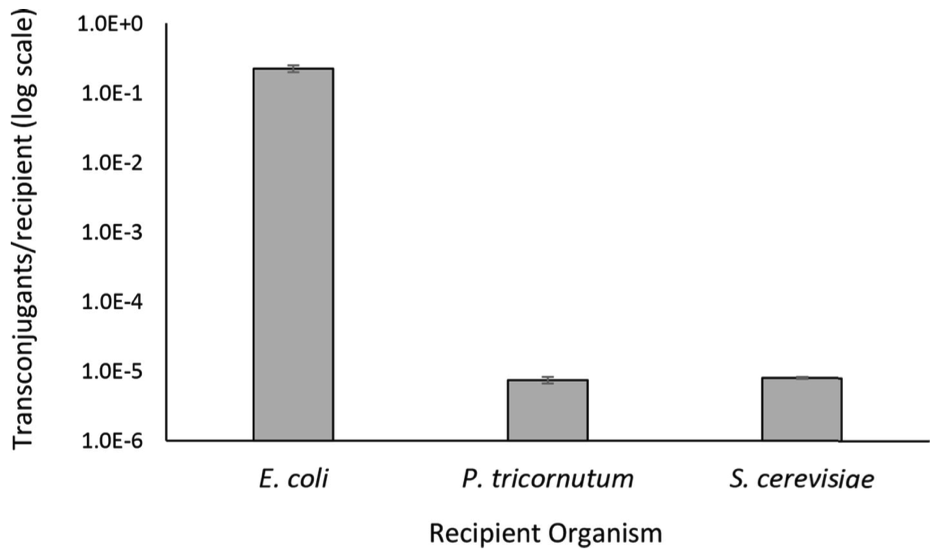 Figure 3. Conjugation efficiency of direct transfer of pAGE1.0 from S. meliloti donor to E. coli, S. cerevisiae, and P. tricornutum recipients. Conjugation efficiency for each donor-recipient pair. Post-conjugation non-selective plates and pAGE1.0 selective plates were used to determine the conjugation efficiency, calculated as transconjugants/ recipient, between each donor and recipient pair. Efficiencies are represented as bar plots with three biological and three technical replicates, with error bars illustrating standard error of the mean.
Figure 3. Conjugation efficiency of direct transfer of pAGE1.0 from S. meliloti donor to E. coli, S. cerevisiae, and P. tricornutum recipients. Conjugation efficiency for each donor-recipient pair. Post-conjugation non-selective plates and pAGE1.0 selective plates were used to determine the conjugation efficiency, calculated as transconjugants/ recipient, between each donor and recipient pair. Efficiencies are represented as bar plots with three biological and three technical replicates, with error bars illustrating standard error of the mean.
Recommended citation: SL. Brumwell, MR. MacLeod, T. Huang, RR. Cochrane, RS. Meaney, M. Zamani, O. Matysiakiewicz, KN. Dan, P. Janakirama, DR. Edgell, TC. Charles, TM. Finan, BJ. Karas. (2019). “Designer Sinorhizobium meliloti strains and multi-functional vectors enable direct inter-kingdom DNA transfer.” PLoS ONE. 14(7): e0219562.
